R Markdown
Alexia Cardona
RStudio
We will be using RStudio both for this R Markdown session and also during the GitHub session. RStudio is currently a very popular Integrated Development Environment (IDE) for working with R. An IDE is an application used by software developers that faciliates programming by offering source code editing, building and debugging tools all integarated into one application. To function correctly, RStudio needs R and therefore both need to be installed on your computer.
The RStudio Desktop open-source product is free under the Affero General Public License (AGPL) v3. Other versions of RStudio are also available.
We will use RStudio IDE to write code, navigate the files on our computer, inspect the variables we are going to create, and visualize the plots, tables and the notebooks that we will generate. RStudio can also be used for version control and we will be looking at this later on.
RStudio interface screenshot. Clockwise from top left: Source, Environment/History, Files/Plots/Packages/Help/Viewer, Console.
RStudio is divided into 4 “Panes”: the Source for your scripts and documents (top-left, in the default layout), your Environment/History/Build/Git (top-right), your Files/Plots/Packages/Help/Viewer (bottom-right), and the R Console (bottom-left). The placement of these panes and their content can be customized (see menu, Tools -> Global Options -> Pane Layout).
One of the advantages of using RStudio is that all the information you need to write code is available in a single window. Additionally, with many shortcuts, autocompletion, and highlighting for the major file types you use while developing in R, RStudio will make typing easier and less error-prone.
Creating an R Markdown project
Before starting to write code in RStudio, we need to create an R Project. The idea behind an R project is to have a workspace where you can keep all the files and settings associated with the project together. In that way, next time you open the R Project it would be easier to resume work. To create an R notebook project for the web in RStudio follow the following steps:
File->New Project->New Directory->Simple Rmarkdown website.- Specify where you would like to create your project. This would be
your working directory.
- Click
Create Project.
RStudio automatically generates a template for you to develop
further. To see how this template would look as a website, click on the
Build tab on the top right hand side pane of RStudio and
then click Build Website.
RStudio’s default preferences generally work well, but saving a
workspace to .RData can be cumbersome, especially if you
are working with larger datasets as this would save all the data that is
loaded into R into the .RData file.
To turn that off, go to Tools ->
Global Options and select the ‘Never’ option for
Save workspace to .RData' on exit.
Set ‘Save workspace to .RData on exit’ to ‘Never’
Working directory
Whenever we are working on a project in RStudio, it is good practice to keep a set of related files e.g., data, images, and code, self-contained in a single folder, called the working directory. All of the scripts within this folder can then use relative paths to files in the working directory that indicate where inside the project a file is located (as opposed to absolute paths, which point to where a file is on a specific computer). Working this way makes it a lot easier to move your project around on your computer and share it with others without worrying about whether or not the underlying scripts will still work.
Absolute vs Relative paths examples
Relative path: data/dataset1.txt
Absolute path: C:/Users/User1/Documents/R/my-first-project/data/dataset1.txt
Using RStudio projects makes it easy to organise your files in the
project and ensures that your working directory is set properly. RStudio
shows your current working directory at the top of your window:
Another way to check your working directory is by typing
getwd() in the console pane. If for some reason your
working directory is not what it should be, you can change it in the
RStudio interface by navigating in the file browser where your working
directory should be, and clicking on the blue gear icon
More, and select Set As Working Directory.
Alternatively you can type
setwd("/path/to/working/directory") in the console to reset
your working directory (not recommended). However, your scripts should
not include this line because it will fail on someone else’s
computer.
Using a consistent folder structure across your projects will help keep things organized, and will also make it easy to find things in the future. This can be especially helpful when you have multiple projects. In general, you may create directories (folders) for data, and images.
data/Use this folder to store your raw data and intermediate datasets you may create for the need of a particular analysis. For the sake of transparency and provenance, you should always keep a copy of your raw data accessible and do as much of your data cleanup and preprocessing programmatically (i.e., with scripts, rather than manually). Separating raw data from processed data is also a good idea. For example, you could have filesdata/raw/survey.plot1.txtanddata/raw/survey.plot2.txtkept separate from adata_output/survey.csvfile generated by thescripts/01.preprocess.survey.Rscript.img/This would be a place to keep figures that you would like to display in your notebook.scripts/This would be the location to keep your R scripts for different analyses or plotting.
You may want additional directories or subdirectories depending on your project needs, but these should form the backbone of your working directory.
The working directory is an important concept to understand. It is the place from where R will be looking for and saving the files. When you write code for your project, it should refer to files in relation to the root of your working directory and only need files within this structure.
R Markdown
An R Markdown file is made up of 3 basic components:
- header
- markdown
- R code chunks
In this course we will assume that the output of our report is an .html file. HTML files are files for web pages. This means that the report that we will generate can be easily deployed on the web.
Header
The markdown document starts with an optional header in YAML format known as the YAML metadata. In the example below the title, author and date are specified in the header. Other options can be specified in the header such as table of content which we will look at later on in the course.
---
title: "My first notebook"
author: Alexia Cardona
date: 18 February 2020
---Hint
Press the
button to see how the report will look like after you make changes to the .Rmd file.
Markdown
The text following the header in an R Markdown file is in Markdown syntax. This is the syntax that gets converted to HTML format once we click on the Knit button or the Build website button. The philosophy behind Markdown is that it should be easy to write and easy to read.
The full documentation of the Markdown syntax can be found at https://pandoc.org/MANUAL.html. However this can be a bit difficult to follow for beginners, therefore below is a simplified version of the Markdown syntax.
Headings
Below is the Markdown code you need to use to specify
headings at different levels and the rendered output respectively below
the code:
# Heading 1
Heading 1
## Heading 2
Heading 2
### Heading 3
Heading 3
#### Heading 4
Heading 4
Inline text formatting
To make text bold use
**double asterisks** or
__double underscores__.
To make text italic use *asterisks* or
_underscores_.
To make text superscript use ^caret^.
To make text subscript use ~tilde~.
To mark text as inline code use
`backticks`.
To strikethrough text use ~~double tilde~~.
Line breaks
To create a line break, put more than 2 spaces at the end of a
sentence or place \ in a new line followed by a new line
.
Challenge
Replicate the output of the text below in R Markdown. The text comes from a paper by Monya Baker that was published in 2016 in the Nature journal that triggered the discussion about Reproducibility Crisis.
Show Answer
--- title: "1,500 scientists lift the lid on reproducibility" author: Monya Baker date: 25 May 2016 --- \ ## Survey sheds light on the 'crisis' rocking research. **More than 70% of researchers have tried and failed to reproduce another scientist's experiments**, and more than half have failed to reproduce their own experiments. Those are some of the telling figures that emerged from *Nature*'s survey of 1,576 researchers who took a brief online questionnaire on reproducibility in research. The data reveal sometimes-contradictory attitudes towards reproducibility. Although 52% of those surveyed agree that there is a significant 'crisis' of reproducibility, less than 31% think that failure to reproduce published results means that the result is probably wrong, and most say that they still trust the published literature.
Links
Linking text to Headers
To link text to a header you would need to specify an identification tag next to a header as follows:
# Markdown {#markdown-header}
Then to link text to this header use
[link to header](#markdown-header). This will be rendered
as link to header.
Linking text to a webpage
To create a link to a webpage use
[text of link](https://training.cam.ac.uk/bioinformatics/event-timetable).
This is rendered as text of
link.
Footnotes
To indicate a footnote use[^1] and, for example,
indicate another one as[^2] and then specify the wordings
of the footnotes as:
[^1]: This is the first footnote.
[^2]: This is the second footnote.
You do not need to put footnotes at the end of the document for them to be rendered there. This example is rendered as follows:
To indicate a footnote use1 and, for example, indicate another one as2 and then specify the wordings of the footnotes as:
Lists
Ordered lists
To create an ordered list use the following syntax:
1. Item 1
2. Item 2
3. Item 3This is rendered as:
- Item 1
- Item 2
- Item 3
Use 4 spaces to indent an item if you would like to have sub-lists:
1. Item 1
2. Item 2
3. Item 3
a. Item 3a
i. Item 3ai
ii. Item 3aii
b. Item 3b
c. Item 3c
4. Item 4- Item 1
- Item 2
- Item 3
- Item 3a
- Item 3ai
- Item 3aii
- Item 3b
- Item 3c
- Item 3a
- Item 4
Unordered lists
In an unordered bulletted list, each item begins with *,
+ or -. Example:
* Item 1
* Item 2
* Item 3
* Item 3a
* Item 3ai
* Item 3aii
* Item 3b
* Item 3c
* Item 4Will be rendered as:
- Item 1
- Item 2
- Item 3
- Item 3a
- Item 3ai
- Item 3aii
- Item 3b
- Item 3c
- Item 3a
- Item 4
Tasks list
Tasks list can be done using the following syntax:
- [ ] an unchecked task list item
- [x] checked itemThis will be rendered as:
- an unchecked task list item
- checked item
Inserting images
To insert an image use the following syntax:
. Example:

Will be rendered as:
R Logo
Challenge
Continue working on the example we created in Challenge 1 to extend it to an output that is similar to this.Show Answer
--- title: "1,500 scientists lift the lid on reproducibility" author: Monya Baker date: 25 May 2016 --- \ ## Reproducibilty Crisis **More than 70% of researchers have tried and failed to reproduce another scientist's experiments**, and more than half have failed to reproduce their own experiments. Those are some of the telling figures that emerged from *Nature*'s survey of 1,576 researchers who took a brief online questionnaire on reproducibility in research. The data reveal sometimes-contradictory attitudes towards reproducibility. Although 52% of those surveyed agree that there is a significant 'crisis' of reproducibility, less than 31% think that failure to reproduce published results means that the result is probably wrong, and most say that they still trust the published literature. 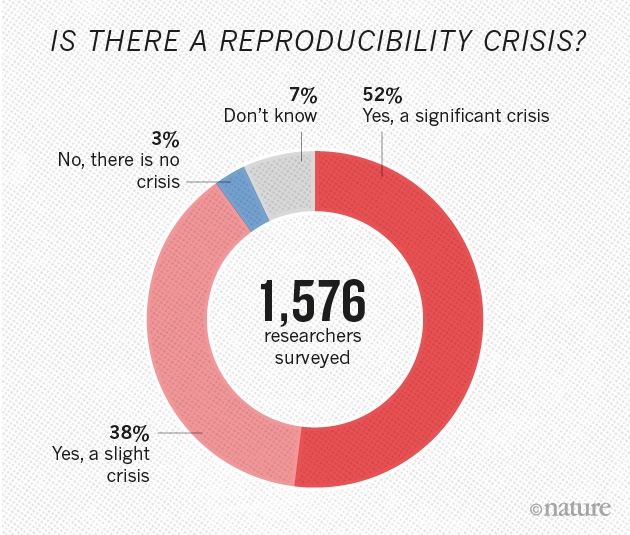 \ ### The Survey In the survey [^1] respondents were asked to rate 11 different approaches to improving reproducibilty in science. Below is the list order by the most highly rated: * Better understanding of statistics * Better mentoring/supervision * More robust design * Better teaching * More within-lab validation * Incentives for better practice * Incentives for formal reproduction * More external-lab validation * More time for mentoring * Journals enforcing standards * More time checking notebooks [^1]: The survey can be downloaded [here](https://www.nature.com/news/polopoly_fs/7.36741!/file/Reproduciblility%20Questionnaire.doc).
Tables
Use |and - to create a table as
follows:
| Column 1 | Column 2 |
| ----------- | ----------- |
| Item 1,1 | Item 1,2 |
| Item 2,1 | Item 2,2 |This is rendered as:
| Column 1 | Column 2 |
|---|---|
| Item 1,1 | Item 1,2 |
| Item 2,1 | Item 2,2 |
Table alignments can be done using the following syntax:
| Left align | Center align | Right align |
| :--- | :----: | ---: |
| Item 1,1 | Item 1,2 | Item 1,3 |
| Item 2,1 | Item 2,2 | Item 2,3 |This is rendered as:
| Left align | Center align | Right align |
|---|---|---|
| Item 1,1 | Item 1,2 | Item 1,3 |
| Item 2,1 | Item 2,2 | Item 2,3 |
Blocks
Blocks in the notebook can be created by using the >
sign as follows: > Example of a block
This is rendered as:
Example of a block
If you would like to add code blocks, use
``` before and after the code as follows:
```
print("Hello world")
x <- 1+2
print(x)
```This will be rendered as:
print("Hello world")
x <- 1+2
print(x)
Adding a table of contents
To add a table of contents to your report add the following to the YAML header:
output:
html_document:
toc: true
toc_float: trueBy default all headings up to level 3 headings are displayed in the
table of contents. You can adjust this by specifying
toc_depth as following:
output:
html_document:
toc: true
toc_depth: 4
toc_float: true
(Optional) Adding references
Adding references and citations in Markdown is not as easy as
reference manager software such as Mendeley. To be able to create
citations you will need to create a bibliography file with all the
references in it. Here is an example of a
bibliography file. The bibligraphy file has to be placed in the same
folder as the one where the .Rmd file is. Next, add the
following to the YAML header:
bibliography: references.bib
link-citations: yesYour YAML header should now look like:
---
title: "My first notebook"
author: Alexia Cardona
date: 18 February 2020
bibliography: references.bib
link-citations: yes
---To cite a reference use the @ together with the ID of
the reference. Example:
Citation to my paper @cardona2014 and @cardona2019
Will be rendered as:
Citation to my paper L. A. A. Cardona Alexia AND Pagani (2014) and A. Cardona et al. (2019)
To add the bibliography at the end of the report add a References heading at the end of the report:
# References
See references at the end of the report.
See https://rmarkdown.rstudio.com/authoring_bibliographies_and_citations.html for more information.
Inserting R Code in Markdown
So far we have not used any R code and all the code we used so far is
in Markdown. As the name suggests, R Markdown files contain Markdown and
R. R Markdown files have a .Rmd extension. Using R in a
Markdown document is useful if we want dynamically generated information
such as integrating our analysis in our report. Some operations that we
might need to do are; loading our dataset, performing some operations on
the dataset and displaying results, either in a table or in a plot. We
will be exploring how we can integrate R code in R Markdown in the
following worked example.
- Create a new R Markdown document (
File->New File->R Markdown)- The file created should already contain some R Markdown code and it should look like this:
- Delete the existing code leaving only the R Markdown header so that we can start building our code as we go over the worked example.
- Name your R Markdown document as RNotebookExample.Rmd
The dataset
Our worked example uses data from gapminder. Let us download the
gapminder_data.csv dataset into our project. Create a
data folder in your working directory. Download the dataset
into the data folder as follows:
download.file(url="https://raw.githubusercontent.com/cambiotraining/reproducibility-training/master/data/gapminder_data.csv", destfile="data/gapminder_data.csv")The gapminder_data.csv dataset that is loaded into the
notebook contains the life expectancy and GDP per capita data for each
country per year from 1952 to 2007. The dataset has the following
columns:
| Column | Description |
|---|---|
| country | Country |
| year | Year the life expectency and GDP per capita index applies |
| pop | Population size |
| continent | Continent |
| lifeExp | Life expectancy |
| gdpPercap | GDP per capita index |
The dataset contains data for different countries in the world. For our analysis we would like to look at only the European countries in the dataset. This means that we would need to manipulate the dataset before we can display the results.
R code chunks
To be able to insert R code inside a notebook you will have to insert
it inside an R code chunk to be able to execute it. To do this either
click on the Insert button at the top of the Source panel
in RStudio:
This creates an R code chunk as follows:
Alternatively, you can type the r code chunk. The R code should be placed in between the triple backticks. Note that on the right hand side of the R code chunk there is a green “play” button that will run the R code chunk if pressed. By default, when the R Markdown document is knitted, the R code will be executed and the R code chunk is displayed before the executed code.
Chunk options
As mentioned, by default the chunk output will be displayed exactly
after the R code chunk and the R code chunk is also displayed in the
rendered page. If you would like to have different settings, you can
specify these as arguments in the {} part of the R code
chunk.
These chunk options include:
include = FALSEdo not display the code and results in the page after it is knitted. The R code however still runs and therefore the variables or results in this code chunk can be used by the other chunks.
echo = FALSEdoes not display the code, but it displays the results in the rendered file.
message = FALSEdoes not display any messages that are generated by the code chunk in the rendered file.warning = FALSEdoes not display warnings that are generated by code chunk in the rendered file.
A full list of options that can be used in the R code chunks is found here.
Chunk label
Most likely you will specify several R code chunks in your R Markdown file. It is recommended practice to label each R code chunk. This will also help navigating your code easily. At the bottom right of your RStudio you should see the code navigator where you can navigate the different code chunks:

To label your chunk just add a label next to r in your R
code chunk as follows:
```{r label-name}
```For our worked example, create an R code chunk and name it as
load-data and add the following code into your first
chunk:
```{r load-data}
#load tidyverse library
library(tidyverse) # used for data manipulation
library(rmarkdown) # used for paged_table function
library(kableExtra) # used for table
library(ggpubr) #used for ggarrange function
#read file into R
pop_data <- read_csv("data/gapminder_data.csv")
#create a table with data from European countries in 2007 showing the countries
#with the largest life expectancy at the top
euro_data_tbl <- pop_data %>%
filter(continent == "Europe" & year == 2007) %>%
select(-continent, -year) %>%
arrange(desc(lifeExp)) %>%
rename(Country = country, "Population Size" = pop,
"Life Expectancy" = lifeExp, "GDP" = gdpPercap)
```We will look at some functions present in the rmarkdown
and kableExtra packages later on. Knit the
RNotebookExample.Rmd file and see what happens. You may
notice that both the code and the output that you would expect to be
printed in the console when you run the code are printed in the
notebook. We would like to have only the code printed in the notebook.
To do that we need to specify the message=FALSE as the
chunk option for our load-data chunk as discussed
previously in the chunk options
section.
```{r load-data, message=FALSE}If you knit RNotebookExample.Rmd again you will see that
now only the code is being printed, which is what we wanted. In the
load-data R code chunk, we have loaded the packages we will
be using in our report and also the data we will be working with. We
then filtered our dataset to contain data from only European countries
in 2007 ordered by life expectancy. The next step is to display this
filtered dataset that is saved in the variable eur_data_tbl
in a table.
Tables in R Markdown
When you have a large dataset, creating a table manually in R Markdown as we did here is not really feasible. Also if you are really trying to make your research reproducible, then, you will need to generate most of the tables dynamically and therefore you will not be able to manually type them in markdown. This is where R Markdown comes in as there is a way to display tables dynamically. In this course we will look at two ways of displaying tables dynamically, using the functions:
kbl()paged_table()
kbl()
The function kbl() is present in the package
kableExtra, a popular package to display tables in R
Markdown. There is extensive documentation on how you can use the
functions within the kableExtra package to draw tables here
which has good examples on how to change the different settings of a
table, such as font, width, highlights, etc…
As a first step, to draw a table you need to use the function
kbl(). The kbl() function takes as an input
the dataset you want to display in a table, e.g.,
kbl(euro_data_tbl).
We have already loaded the package kableExtra in the
load-data R code chunk. Create a new R code chunk called
kbl-table and draw the a table with the
eur_data_tbl tibble.
```{r kbl}
euro_data_tbl %>%
kbl()
```Let us add a style to the table so that it looks more attractive. The
kable_styling() function is the function used to customise
the way the table looks. We are going to add the striped
style so that the table have striped rows and also the
hover style so that when you move your mouse over the table
you can see it hover.
```{r kbl}
euro_data_tbl %>%
kbl() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
```
Challenge
Our table is getting prettier. Try a few styles with
kable_styling(). Look at the documentation for ideas. After you have tried a few styles, set your R Markdown file to look like thisExperiment using the different chunk options and see what the output will be link with each option. Refer to chunk options section.
paged_table()
If you have a very long table to display, the best way would be to
use pagination where the contents of the table are split into multiple
tabs. The function paged_table in the
rmarkdown library can be used to do this. It is also very
simple to use.
Create a new R code chunk called paged-table and use the
paged_table() function to display the euro_data_tbl table
as follows:
```{r paged-table}
paged_table(euro_data_tbl)
```
Adding images
Adding images is straightforward and does not require using a specific R Markdown function.
Challenge
Create a new dataset
euro_data_figby filtering thepop_datatibble to contain only data from Europe. Draw a plot to display thelifeExpon the y axis andyearon the x axis. Usegeom_violin()to draw this as a violin plot to show the distrubution of the data across each year and save it in aeuro_plotvariable.
Optional
In reports or publications, sometimes you would need to place two or
more figures next to each other. ggarrage() is a function
present in the ggpubr pacakge that can help with this. The
ggarrange() function takes as an input the list of plots to
plot. You can specify the number of columns and rows to plot by the ncol
and nrow arguments respectively. Another thing that you might need is a
lable for each plot so that you can specify the description in your
caption. Use the labels argument to specify labels.
Challenge
Create a new dataset
uk_data_figby filtering thepop_datatibble to contain only data from the United Kingdom. Draw a scatter plot to display thelifeExpon the y axis andyearon the x axis and save it in auk_plotvariable.
Draw theeuro_plotcreated in the previous challenge next to auk_plotusing theggarrange()function. Label the plots A and B respectively.
Summary
Through R notebooks we can now integrate our R code and our results in one readable document that we can share with our collaborators. In this way the code and the results have become one item. For inspiration, examples of R notebooks can be found at https://rpubs.com/